 |
 |
|
|
| Modular Strategy for Tailoring Functional Ribonucleopeptides |
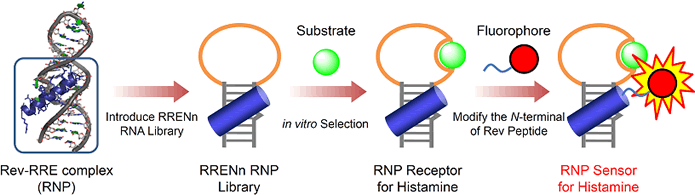
A stable complex of a peptide and RNA, ribonucleopeptide (RNP), provides
a new framework to construct a macromolecular receptor for small molecules.
The RRE (Rev Responsive Element) RNA and the Rev peptide form a structurally
well-characterized stable RNP complex that is suitable for molding of a
ligand-binding pocket of the RNP in a step-wise manner.
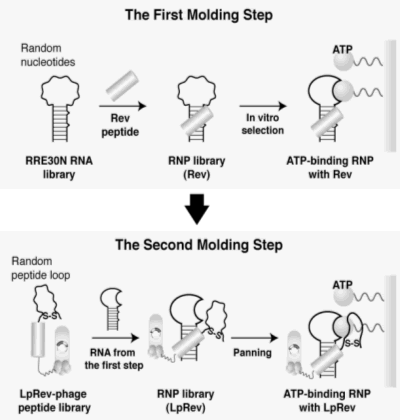
The first step involves molding of the RNA subunit by in vitro selection of
an RNP pool originating from an RNA library and the Rev peptide. The second step involves selection
from an RNP library consisting of Rev peptides with randomized amino-acid residues and the RNA subunit
selected in the first molding. Together, these steps afford an expanded ligand-binding surface
consisting of both RNA and peptide subunits. We report here a step-wise molding strategy to tailor
an ATP-binding pocket in the RNP. The ATP-binding pocket produced by sequential molding of RNA and
peptide subunits shows higher affinity and specificity to ATP than the ATP-binding RNP receptor
in which only the RNA subunit was molded. In vitro selection of the RNP pool originating from an
RRE-based RNA library and the Rev peptide affords RNP receptors specific for nucleotide triphosphates
or the phosphotyrosine residue.
The peptide-based molding of the ATP-binding RNP was carried out next by using a
peptide-derived RNP library constructed by the phage display technique along with an RNA subunit of
the ATP-binding RNP obtained from the first RNA-based library selection. An LpRev peptide was designed
to possess seven randomized amino-acids (Cys-X7-Cys) at the N-terminus of Rev to facilitate efficient
formation of a binding surface. The random LpRev peptides were expressed at the amino terminus of the
pIII protein of the filamentous bacteriophage, and expected to form a loop structure upon disulfide
bond formation. Phages from the LpRev library were pooled and mixed with the RNA obtained in the first
molding step to form a library of RNPs before starting selection against ATP-agarose resin.
One of the RNP bearing the LpRev peptide shows higher affinity and selectivity to ATP over dATP than the parent ATP-binding RNP. |
 |
 |
|
Figure 1. A schematic illustration shows the step-wise molding strategy to generate ribonucleopeptide (RNP) receptors for ATP.
The first molding step utilizes in vitro selection of an RNP library originating from a randomized RNA nucleotide pool (RRE30N).
Combination of the RNA subunit of RNP selected in the first molding and a phage peptide library of the Rev peptide containing
seven randomized amino-acid residues affords a peptide-diverged RNP library, which enables the selection in the second molding
step of improved ATP-binding RNPs.
|
 |
| The RNP receptor functionalized by a fluorophore-labeled Rev peptide exerts
optical signals associated with the ligand binding events. Replacing the
Rev peptide of the ATP-binding RNP with a fluorophore-modified Rev peptide
affords a series of fluorescent ATP sensors. This strategy to generate
tailor-made fluorescent sensors is applied for a selective detection of
a specific phosphorylated tyrosine residue within a defined amino acid
sequence. |
 |
| A new strategy for tailoring fluorescent biosensors. |
 |
| Fluorescent biosensors that facilitate reagentless sensitive detection
of small molecules are crucial tools in the areas of therapeutics and diagnostics.
However, construction of fluorescent biosensors with desired characteristics,
i.e., detection wavelengths and concentration ranges for ligand detection,
from macromolecular receptors is not a straightforward task. An ATP-binding
RNP receptor was converted to a fluorescent ATP sensor without chemically
modifying the nucleotide in the ATP-binding RNA. The RNA subunit of the
ATP-binding RNP and a peptide modified with a pyrenyl group formed a stable
fluorescent RNP complex that showed an increase in the fluorescence intensity
upon binding to ATP.
|
 |
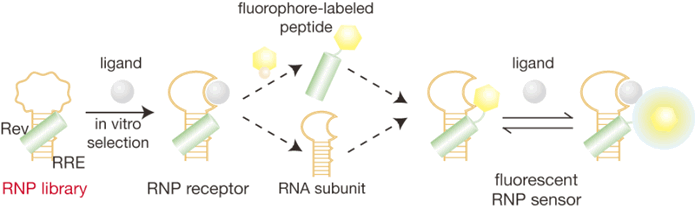 |
| Figure 2. A scheme illustrates a direct conversion of a ribonucleopeptide
(RNP) receptor to a fluorescent RNP sensor. A complex of the Rev peptide
and RRE RNA was used as a framework for the RNP receptor and RNP sensor. |
 |
A simple screening scheme based on the relative changes in emission intensities
of fluorescent RNPs with or without the ligand ATP (I/I0) is quite useful for the screen of RNP sensors with desired emission wavelength
and with high I/I0 ratio with emission wavelengths varying from 390 to 670 nm. Screening
of the fluorescence emission intensity changes in the presence of increasing
concentrations of ATP allowed titration analysis of the fluorescent RNP
library, which provided ATP sensors responding at wide concentration ranges
of ATP. Simultaneous application of these four sensors covers ATP concentration
ranges from ~10-7 to ~10-2 M.
The combinatorial strategy using the modular RNP receptor reported here enables tailoring of a fluorescent sensor for a specific ligand without knowledge of detailed structural information for the macromolecular receptor. |
 |
 |
 |
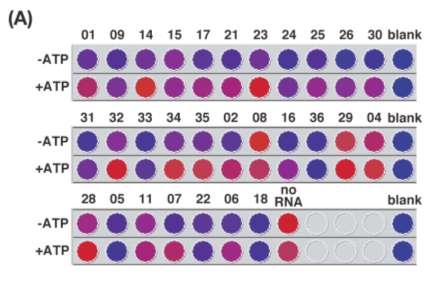
| Figure 3A. A scanned image showing the microplate assay for the combinatorial
screening of the fluorescent RNP library. Fluorescence intensities of 7mC-Rev
derived RNPs (1 μM) were evaluated in the absence or presence of 1 mM of
ATP with excitation at 355 nm and emission 390 nm with intensities being
weak in blue color and strong in red. |
 |
 |
 |
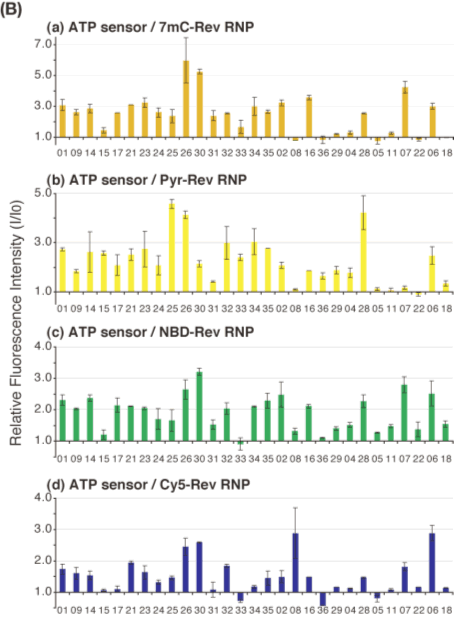
| Figure 3B. Relative fluorescence intensity changes (I/I0) of RNPs upon ATP binding are shown in the bar graphs for (a) 7mC-Rev
RNP, (b) Pyr-Rev RNP, (c) NBD-Rev RNP, and (d) Cy5-Rev RNP. |
 |
 |
 |
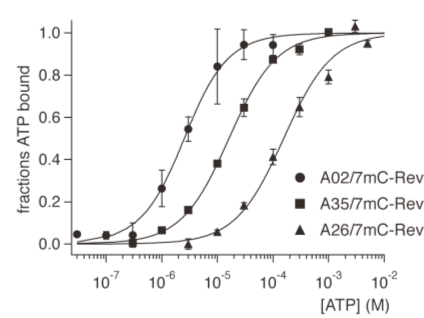
| Figure 4. Saturation curves of 7mC-Rev RNPs A02/7mC-Rev (filles circles), A35/7mC-Rev (filled squares), and A26/7mC-Rev (filled triangles) in the presence of 10 nM to 10 mM ATP were determined by titrations of fluorescence intensity changes. |
 |
| Context-Dependent Fluorescence Detection of a Phosphorylated Tyrosine Residue by a Ribonucleopeptide |
 |
Tools for selective recognition and sensing of specific phosphorylated
tyrosine residues on the protein surface are essential for understanding
signal transduction cascades in the cell. In vitro selection of an RNA-derived
pool of RNP afforded RNP receptors specific for a phosphotyrosine residue
within a defined amino-acid sequence Gly-Tyr-Ser-Arg. The RNP receptor
for the specific phosphotyrosine residue was successfully converted to
a fluorescent RNP sensor for sequence-specific recognition of a phosphorylated
tyrosine. The fluorescent RNP sensor has an ability to function as a specific
fluorescent sensor for the phosphorylated tyrosine residue within a defined
amino-acid sequence in HeLa cell extracts.
|
 |
 |
| Figure 5. Strategy to obtain RNP fluorescent sensors specific for a phosphotyrosine-containing
amino acid sequence GpYSR. Combination of the RNA subunit of the GpYSR-binding
RNP and a fluorophore-modified Rev peptide provided a GpYSR RNP fluorescent
sensor. |
 |
| Development of Fluorescent Ribonucleopeptide Sensors for Histamine |
 |
Biologically active amines, such as dopamine, histamine, and serotonin,
play major roles in the regulation of movement and are implicated in the
pathophysiology of Parkinson’s and Huntington’s diseases, psychosis, and
drug addiction. We developed fluorescent RNP sensors for histamine with
a variety of binding and signal-transducing characteristics. The fluorescent
histamine sensor showed distinct selectivity for histamine over structurally
related histamine analogs, such as imidazole, ethylamine and L-histidine.
|
 |
 |
| Figure 6. A strategy to obtain RNP fluorescent sensors specific for histamine.
Combination of the RNA subunits of the histamine-binding RNP receptor and
a fluorophore-modified Rev peptide provided a histamine RNP fluorescent
sensor. |
 |
| These researches were supported by a Grant-in-aid for Scientific Research from Ministry of Education, Science, Sports and Culture, Japan to T.M. (No. 19021023, and No. 20241051). |
 |
| Structual Aspects for the Recognition of ATP by Ribonucleopeptide Receptors |
 |
Rational design of the functional RNPs would be possible by understanding the structural aspects for the function of RNP receptor. A combination of NMR measurements, enzymatic and chemical mapping, and nucleotide mutation studies of the RNP-adenosine complex show that RNP interacts with the adenine ring of adenosine by forming a U:A:U triple with two invariant U nucleotides (Figure 1b). The observed recognition mode for the adenine ring is very different from those of RNA aptamers for ATP derivatives reported by others previously. The RNP-adenosine complex is folded into a particular structure by formation of the U:A:U triple and a Hoogsteen type A:U base pair (Figure 1b). This recognition mechanism was successfully utilized to convert the substrate-binding specificity of RNP from ATP- to GTP-binding with a C+:G:C triple recognition mode.
|
 |
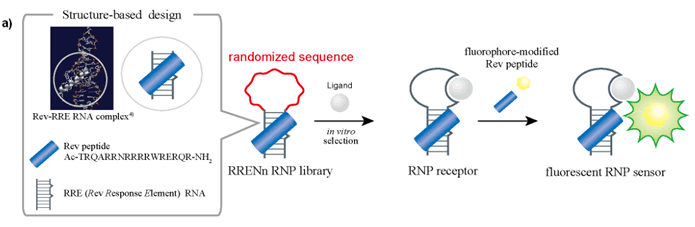 |
| Figure 7A. A scheme shows the stepwise construction of RNP receptors and
fluorescent RNP sensors. |
 |
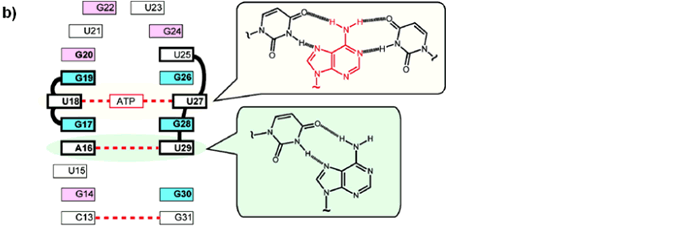 |
| Figure 7B. An illustration shows the proposed recognition mode of ATP-binding
RNP receptor clarified in this study. |
 |
|
|