 |
 |
|
|
 |
| Emerging molecular design principles of functional biomolecules and biomolecular assemblie |
 |
 |
 |
Nature's strategy for efficient energy utilization
A transition to renewable energy technologies requires
new chemistry to learn from nature. Nature has found fantastic solutions
to convert solar energy to produce chemicals and to utilize them in the
exceptionally efficient manners for almost 3 billion years. It is our challenge
to understand the efficient bioenergetic processes of nature and to construct
bio-inspired energy utilization systems.
Our research interests
The research interests in our group focus on the design of biomacromolecules
and their assemblies for molecular recognition, catalysis and signal transduction
in water, the solvent of life. We take synthetic, organic chemical, biochemical
and biophysical approaches to understand the biological molecular recognition
and chemical reactions.
Spacially assembled enzymes: a clue for the efficient metabolic reaction in the cell
While the individual enzyme shows marvelous efficiency, the beauty of the chemical reaction processes
in cells is found in its highly specific multi-step chemical transformation process in the presence of closely related ligands.
Multiple enzymes cooperate to catalyze the sequential steps of metabolic pathway in the efficiency that artificial catalysts still cannot achieve.
Molecular Switchboard
A clue for the efficient sequential metabolic processes
comes from the spatial, and possibly temporal, assembly of multiple enzymes.
To realize such an assembly of enzymes or biomacromolecules, we use a DNA
nanoarchitecture, DNA origami, for a template with defined addresses. Each
unique address on the DNA nanoarchitecture is used to locate an enzyme
of interest to assemble multiple enzyme at defined inter-enzyme distance.
We have developed protein-based adaptors by utilizing the sequence-specific
DNA binding proteins. A protein of interest is fused to the adaptor to
locate functional proteins at the specific sequences on DNA origami to
realize the molecular switchboard.
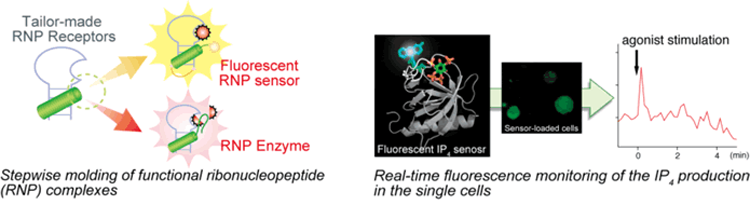
Exploring the function of RNA-peptide (RNP) complexes
Design strategies for receptors, sensors and enzymes are explored by utilizing structurally well-characterized RNA-peptide (RNP) complexes.
Parallel application of the structure-based design and in vitro selection affords highly specific receptors and sensors for biologically important ligands,
such as ATP and the phosphorylated tyrosine residue within a defined amino acid sequence. The information at the biological level, such as the dynamics of
the ligand concentration changes, would be translated into a light signal by the fluorescent biosensors to understand the biological signal transduction mechanisms.
Fluorescent biosensors for cellular second messengers
Small protein domains are explored to realize visualization of cellular signals by fluorescent biosensors,
directed self-assembly of peptides and proteins to build up nanobiomaterials, and reconstitution of the functional assemblies of receptors and enzymes.
A miniature protein derived from the PH domain has been successfully applied for the fluorescent biosensors to real-time monitoring the cellular dynamics of second messengers,
inositol-1,4,5-trisphosphate [Ins(1,4,5)P3] and inositol-1,3,4,5-tetrakisphosphate [Ins(1,3,4,5)P4].
|
 |
Molecular switchboard
|
Artificial
enzyme
|
Biosensor
|
Chemical
Probe
|
 |
|
|